Nick Borcherding
Assistant Professor of Pathology & Immunology
Washington University School of Medicine in St. Louis
Biography
Nick Borcherding is a physician–scientist whose work bridges computational immunology, clinical pathology, and transplant immunogenetics. He serves as an Assistant Professor in the Department of Pathology and Immunology at Washington University School of Medicine, where his clinical practice focuses on human leukocyte antigen testing for transplantation, autoimmunity, and cancer immunotherapy.
Nick’s research explores how the adaptive immune system encodes and recalls disease experiences. His lab integrates single-cell sequencing, systems immunology, and machine learning to map immune diversity and predict clinical outcomes. A particular focus is on the use of innate and adaptive cellular barcodes, including mitochondrial genomes and immune receptor repertoires, to trace clonal relationships across tissues and disease states.
Beyond his laboratory and clinical roles, Nick is deeply invested in open data science. He develops widely used open-source software for immune repertoire and single-cell analysis, including
- Tumor Immunology
- Immunometabolism
- Single-Cell Immune Profiling
- Adaptive Immune Receptor Repertoire Analyses
- Open Data Science
-
Fellow, Histocompatibility & Immunogenetics, Present
Washington University School of Medicine
-
Residency in Clinical Pathology, 2023
Washington University School of Medicine
-
MD / PhD, Cancer Biology, 2020
University of Iowa
-
MS, Pathology, 2014
University of Iowa
-
BS, Nutritional Sciences (Summa Cum Laude), 2012
Iowa State University
Experience
Recent & Upcoming Talks
Featured Publications
Single-cell adaptive immune receptor repertoire sequencing (scAIRR-seq) and single-cell RNA sequencing (scRNA-seq) provide a transformative approach to profiling immune responses at unprecedented resolution across diverse pathophysiologic contexts. This work presents scRepertoire 2, a substantial update to our R package for analyzing and visualizing single-cell immune receptor data. This new version introduces an array of features designed to enhance both the depth and breadth of immune receptor analysis, including improved workflows for clonotype tracking, repertoire diversity metrics, and novel visualization modules that facilitate longitudinal and comparative studies. Additionally, scRepertoire 2 offers seamless integration with contemporary single-cell analysis frameworks like Seurat and SingleCellExperiment, allowing users to conduct end-to-end single-cell immune profiling with transcriptomic data. Performance optimizations in scRepertoire 2 resulted in a 85.1% increase in speed and a 91.9% reduction in memory usage from the first version over the range repertoire size tested in benchmarking, addressing the demands of the ever-increasing size and scale of single-cell studies. This release marks an advancement in single cell immunogenomics, equipping researchers with a robust toolset to uncover immune dynamics in health and disease.
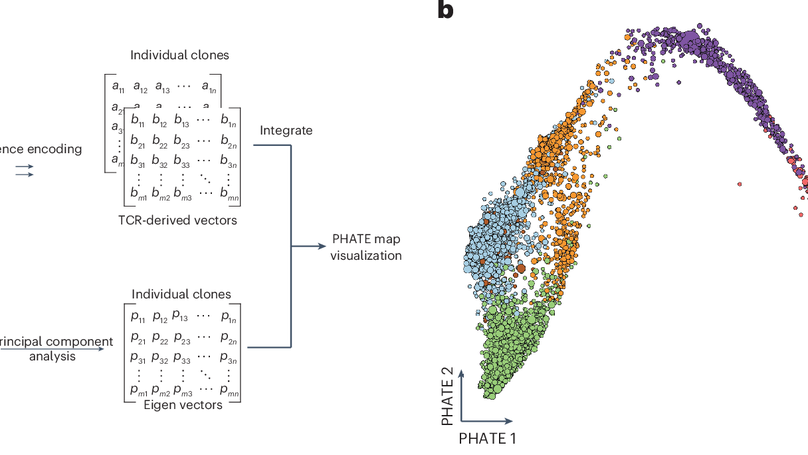
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and mRNA vaccination induce robust CD4+ T cell responses. Using single-cell transcriptomics, here, we evaluated CD4+ T cells specific for the SARS-CoV-2 spike protein in the blood and draining lymph nodes (dLNs) of individuals 3 months and 6 months after vaccination with the BNT162b2 mRNA vaccine. We analyzed 1,277 spike-specific CD4+ T cells, including 238 defined using Trex, a deep learning-based reverse epitope mapping method to predict antigen specificity. Human dLN spike-specific CD4+ follicular helper T (TFH) cells exhibited heterogeneous phenotypes, including germinal center CD4+ TFH cells and CD4+IL-10+ TFH cells. Analysis of an independent cohort of SARS-CoV-2-infected individuals 3 months and 6 months after infection found spike-specific CD4+ T cell profiles in blood that were distinct from those detected in blood 3 months and 6 months after BNT162b2 vaccination. Our findings provide an atlas of human spike-specific CD4+ T cell transcriptional phenotypes in the dLNs and blood following SARS-CoV-2 vaccination or infection.
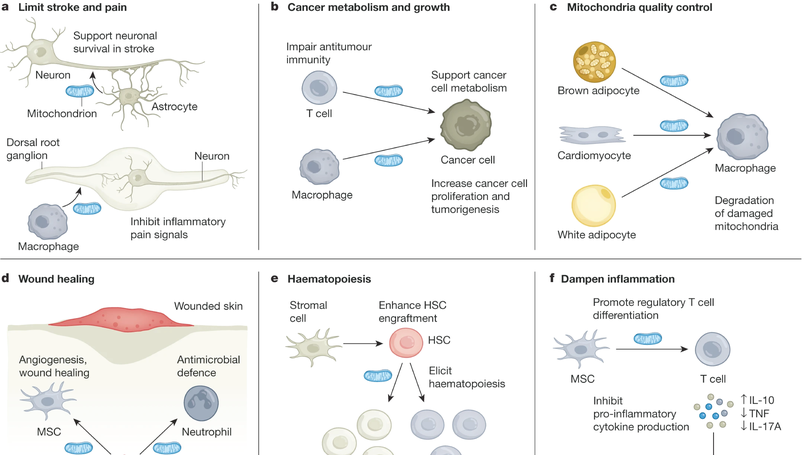
Mitochondria are believed to have originated through an ancient endosymbiotic process in which proteobacteria were captured and co-opted for energy production and cellular metabolism. Mitochondria segregate during cell division and differentiation, with vertical inheritance of mitochondria and the mitochondrial DNA genome from parent to daughter cells. However, an emerging body of literature indicates that some cell types export their mitochondria for delivery to developmentally unrelated cell types, a process called intercellular mitochondria transfer. In this Review, we describe the mechanisms by which mitochondria are transferred between cells and discuss how intercellular mitochondria transfer regulates the physiology and function of various organ systems in health and disease. In particular, we discuss the role of mitochondria transfer in regulating cellular metabolism, cancer, the immune system, maintenance of tissue homeostasis, mitochondrial quality control, wound healing and adipose tissue function. We also highlight the potential of targeting intercellular mitochondria transfer as a therapeutic strategy to treat human diseases and augment cellular therapies.
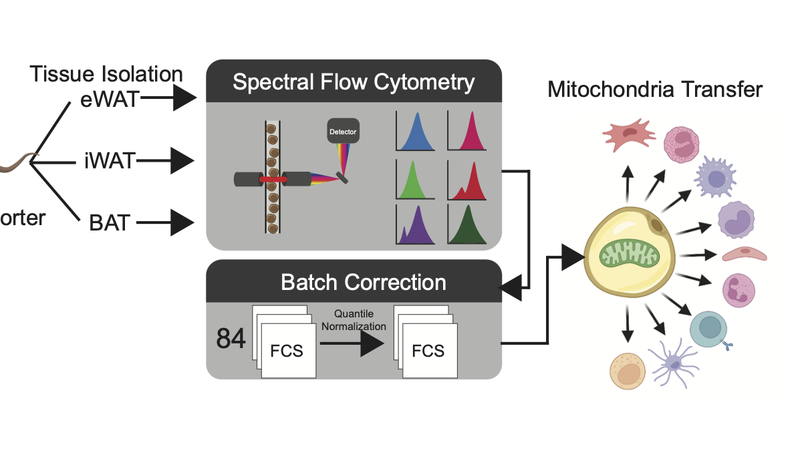
Adipocytes transfer mitochondria to macrophages in white and brown adipose tissues to maintain metabolic homeostasis. In obesity, adipocyte-to-macrophage mitochondria transfer is impaired, and instead, adipocytes release mitochondria into the blood to induce a protective antioxidant response in the heart. We found that adipocyte-to-macrophage mitochondria transfer in white adipose tissue is inhibited in murine obesity elicited by a lard-based high-fat diet, but not a hydrogenated-coconut-oil-based high-fat diet, aging, or a corn-starch diet. The long-chain fatty acids enriched in lard suppress mitochondria capture by macrophages, diverting adipocyte-derived mitochondria into the blood for delivery to other organs, such as the heart. The depletion of macrophages rapidly increased the number of adipocyte-derived mitochondria in the blood. These findings suggest that dietary lipids regulate mitochondria uptake by macrophages locally in white adipose tissue to determine whether adipocyte-derived mitochondria are released into systemic circulation to support the metabolic adaptation of distant organs in response to nutrient stress.
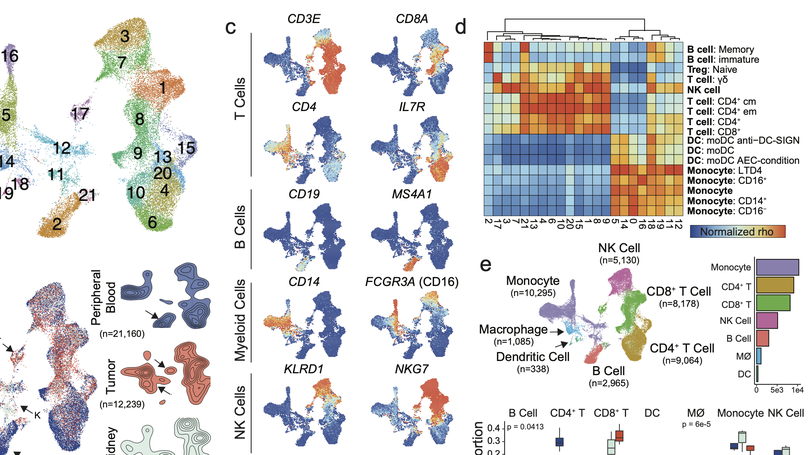
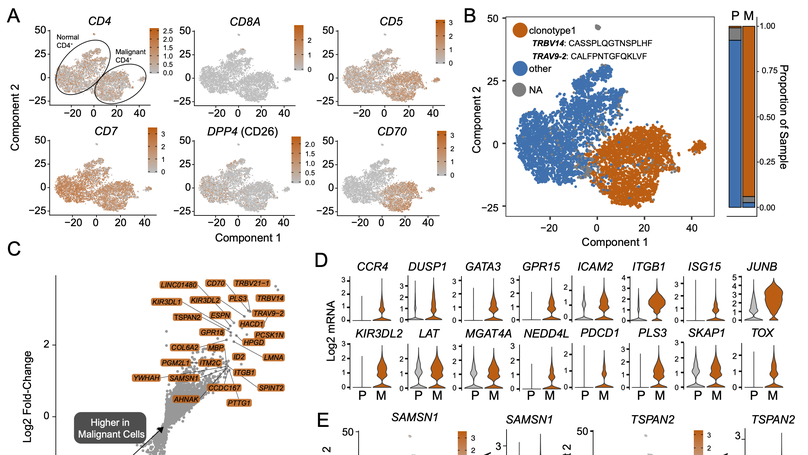
Clear cell renal cell carcinoma (ccRCC) is one of the most immunologically distinct tumor types due to high response rate to immunotherapies, despite low tumor mutational burden. To characterize the tumor immune microenvironment of ccRCC, we applied single-cell-RNA sequencing (SCRS) along with T-cell-receptor (TCR) sequencing to map the transcriptomic heterogeneity of 25,688 individual CD45+ lymphoid and myeloid cells in matched tumor and blood from three patients with ccRCC. We also included 11,367 immune cells from four other individuals derived from the kidney and peripheral blood to facilitate the identification and assessment of ccRCC-specific differences. There is an overall increase in CD8+ T-cell and macrophage populations in tumor-infiltrated immune cells compared to normal renal tissue. We further demonstrate the divergent cell transcriptional states for tumor-infiltrating CD8+ T cells and identify a MKI67 + proliferative subpopulation being a potential culprit for the progression of ccRCC. Using the SCRS gene expression, we found preferential prediction of clinical outcomes and pathological diseases by subcluster assignment. With further characterization and functional validation, our findings may reveal certain subpopulations of immune cells amenable to therapeutic intervention.
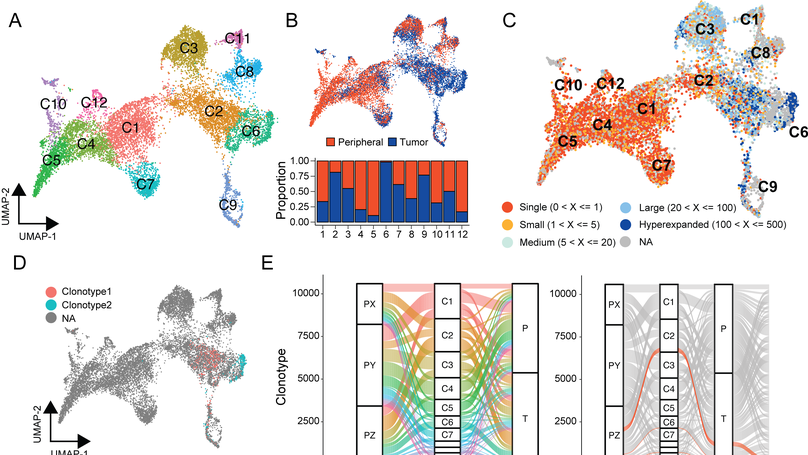
Single-cell sequencing is an emerging technology in the field of immunology and oncology that allows researchers to couple RNA quantification and other modalities, like immune cell receptor profiling at the level of an individual cell. A number of workflows and software packages have been created to process and analyze single-cell transcriptomic data. These packages allow users to take the vast dimensionality of the data generated in single-cell-based experiments and distill the data into novel insights. Unlike the transcriptomic field, there is a lack of options for software that allow for single-cell immune receptor profiling. Enabling users to easily combine mRNA and immune profiling, scRepertoire was built to process data derived from 10x Genomics Chromium Immune Profiling for both T-cell receptor (TCR) and immunoglobulin (Ig) enrichment workflows and subsequently interacts with a number of popular R packages for single-cell expression, such as Seurat. The scRepertoire R package and processed data are open source and available on GitHub and provides in-depth tutorials on the capability of the package.