View the count of clones frequency group in Seurat or SCE object
meta data after combineExpression(). The visualization
will take the new meta data variable cloneSize and
plot the number of cells with each designation using a secondary
variable, like cluster. Credit to the idea goes to Drs. Carmona
and Andreatta and their work with ProjectTIL.
clonalOccupy(
sc.data,
x.axis = "ident",
label = TRUE,
facet.by = NULL,
order.by = NULL,
proportion = FALSE,
na.include = FALSE,
exportTable = FALSE,
palette = "inferno",
...
)Arguments
- sc.data
The single-cell object after
combineExpression()- x.axis
The variable in the meta data to graph along the x.axis.
- label
Include the number of clone in each category by x.axis variable
- facet.by
The column header used for faceting the graph
- order.by
A character vector defining the desired order of elements of the
group.byvariable. Alternatively, usealphanumericto sort groups automatically.- proportion
Convert the stacked bars into relative proportion
- na.include
Visualize NA values or not
- exportTable
If
TRUE, returns a data frame or matrix of the results instead of a plot.- palette
Colors to use in visualization - input any hcl.pals
- ...
Additional arguments passed to the ggplot theme
Value
Stacked bar plot of counts of cells by clone frequency group
Examples
# Getting the combined contigs
combined <- combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L",
"P19B","P19L", "P20B", "P20L"))
# Getting a sample of a Seurat object
scRep_example <- get(data("scRep_example"))
# Using combineExpresion()
scRep_example <- combineExpression(combined, scRep_example)
# Using clonalOccupy
clonalOccupy(scRep_example, x.axis = "ident")
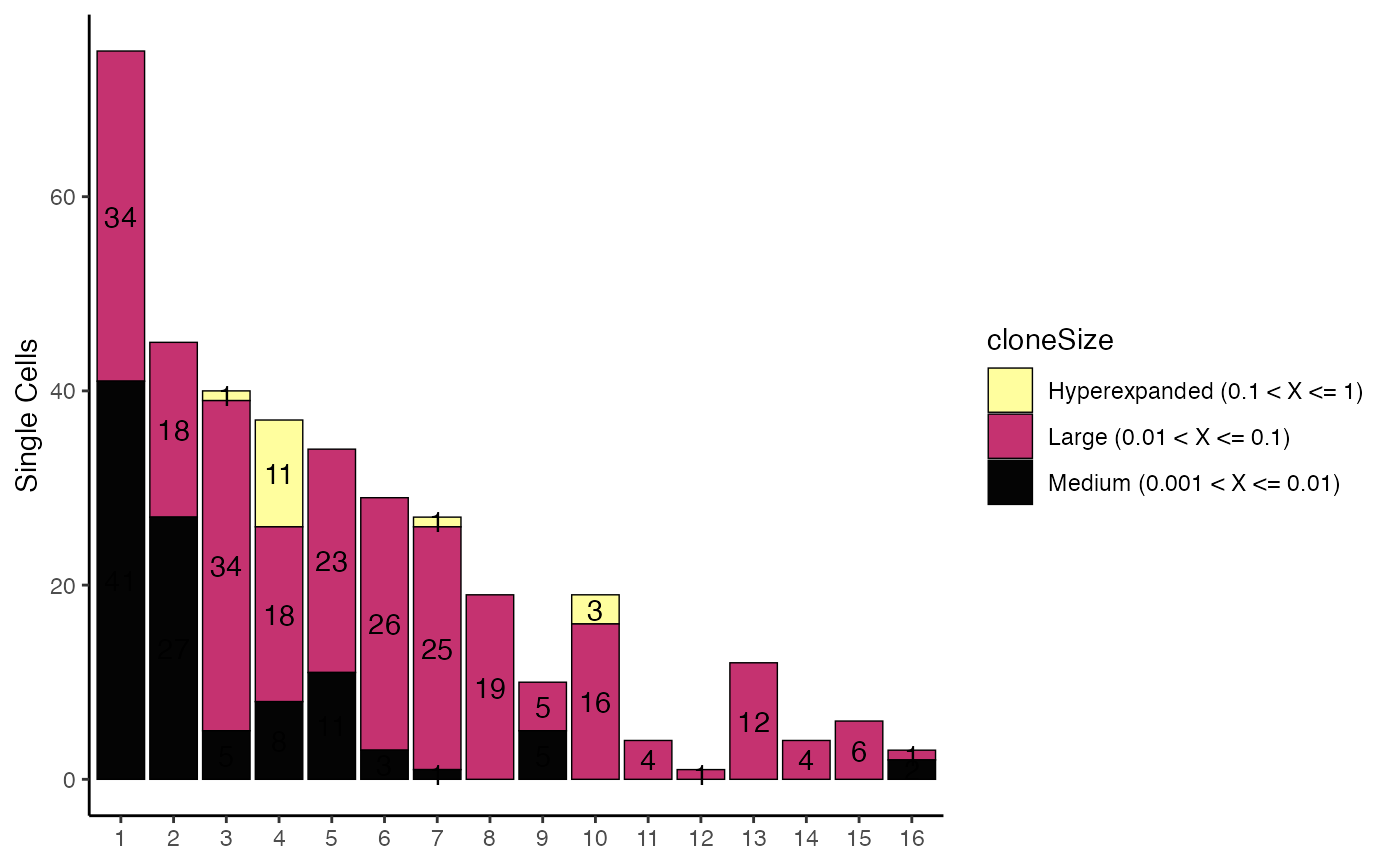 table <- clonalOccupy(scRep_example, x.axis = "ident", exportTable = TRUE)
table <- clonalOccupy(scRep_example, x.axis = "ident", exportTable = TRUE)