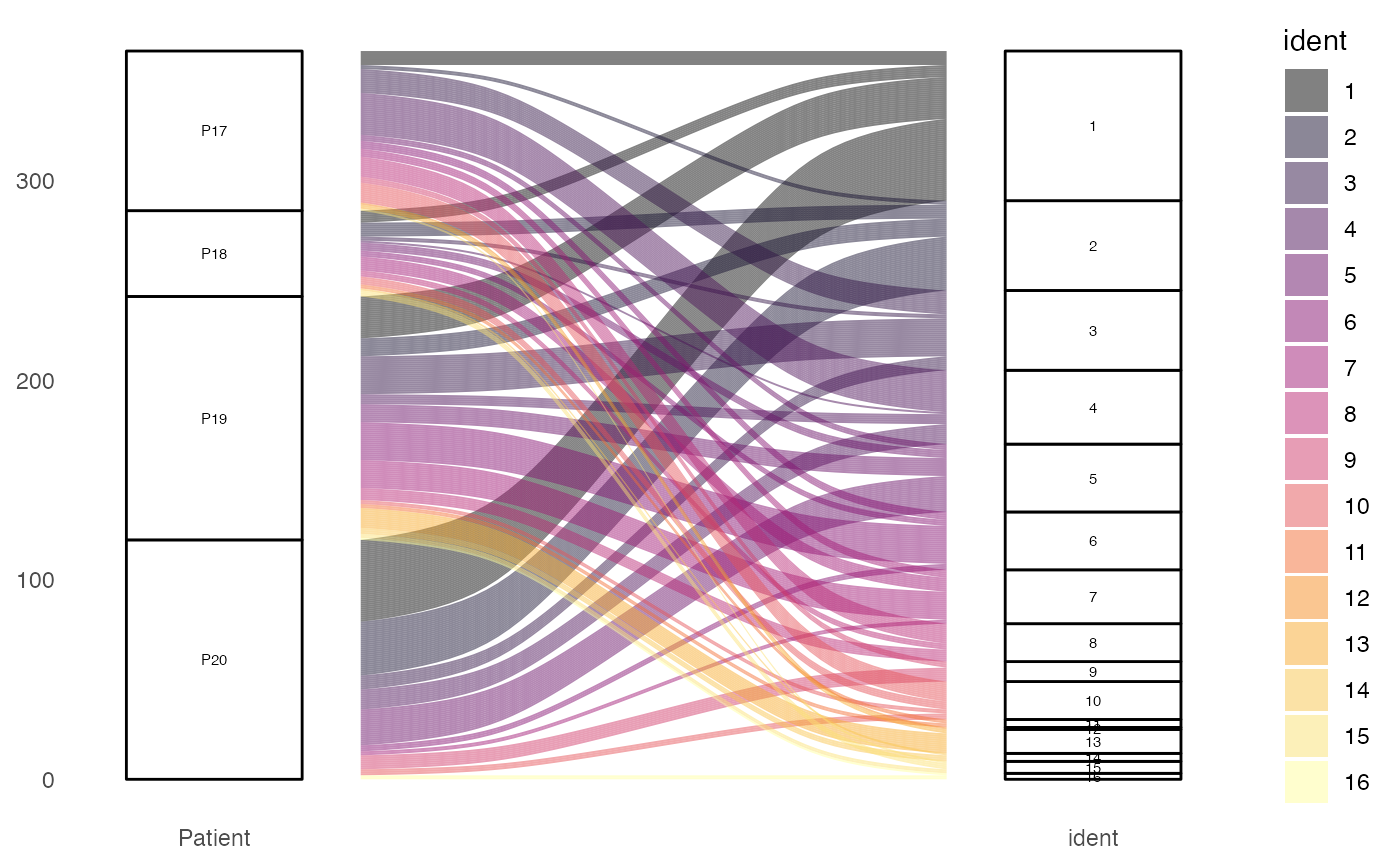
View the proportional contribution of clones by Seurat or SCE object
meta data after combineExpression(). The visualization
is based on the ggalluvial package, which requires the aesthetics
to be part of the axes that are visualized. Therefore, alpha, facet,
and color should be part of the the axes you wish to view or will
add an additional stratum/column to the end of the graph.
alluvialClones(
sc.data,
cloneCall = "strict",
chain = "both",
y.axes = NULL,
color = NULL,
alpha = NULL,
facet = NULL,
exportTable = FALSE,
palette = "inferno",
...
)Arguments
- sc.data
The product of
combineExpression().- cloneCall
Defines the clonal sequence grouping. Accepted values are:
gene(VDJC genes),nt(CDR3 nucleotide sequence),aa(CDR3 amino acid sequence), orstrict(VDJC + nt). A custom column header can also be used.- chain
The TCR/BCR chain to use. Use
bothto include both chains (e.g., TRA/TRB). Accepted values:TRA,TRB,TRG,TRD,IGH,IGL(for both light chains),both.- y.axes
The columns that will separate the proportional . visualizations.
- color
The column header or clone(s) to be highlighted.
- alpha
The column header to have gradated opacity.
- facet
The column label to separate.
- exportTable
If
TRUE, returns a data frame or matrix of the results instead of a plot.- palette
Colors to use in visualization - input any hcl.pals.
- ...
Additional arguments passed to the ggplot theme
Value
A ggplot object visualizing categorical distribution of clones, or a
data.frame if exportTable = TRUE.
Examples
# Getting the combined contigs
combined <- combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L",
"P19B","P19L", "P20B", "P20L"))
# Getting a sample of a Seurat object
scRep_example <- get(data("scRep_example"))
# Using combineExpresion()
scRep_example <- combineExpression(combined, scRep_example)
scRep_example$Patient <- substring(scRep_example$orig.ident, 1,3)
# Using alluvialClones()
alluvialClones(scRep_example,
cloneCall = "gene",
y.axes = c("Patient", "ident"),
color = "ident")
#> Ignoring unknown labels:
#> • width : "0.2"